About
This website makes available the results from Bell C.G. et al., (2017) “Obligatory And Facilitative Allelic Variation In The DNA Methylome Within Common Disease-Associated Loci.”
This analysis was performed in 3,128 MeDIP-seq DNA methylomes derived from peripheral blood. The results present the allelic dosage association of EBI-NIH Catalogue GWAS SNPs with DNA methylation. This was assessed in 500 bp semi-overlapping windows across the respective GWAS SNP Linkage Disequilibrium Blocks. It therefore identifies significant regional DNA methylation variation between risk and non-risk GWAS SNP haplotypes for these robustly associated human diseases and phenotypes. Genetic variation (CNVs, Indels, STRs, etc.) potentially contributing to these signals due to obligatory, facilitative, or dosage effects are subsequently overlaid with these results.
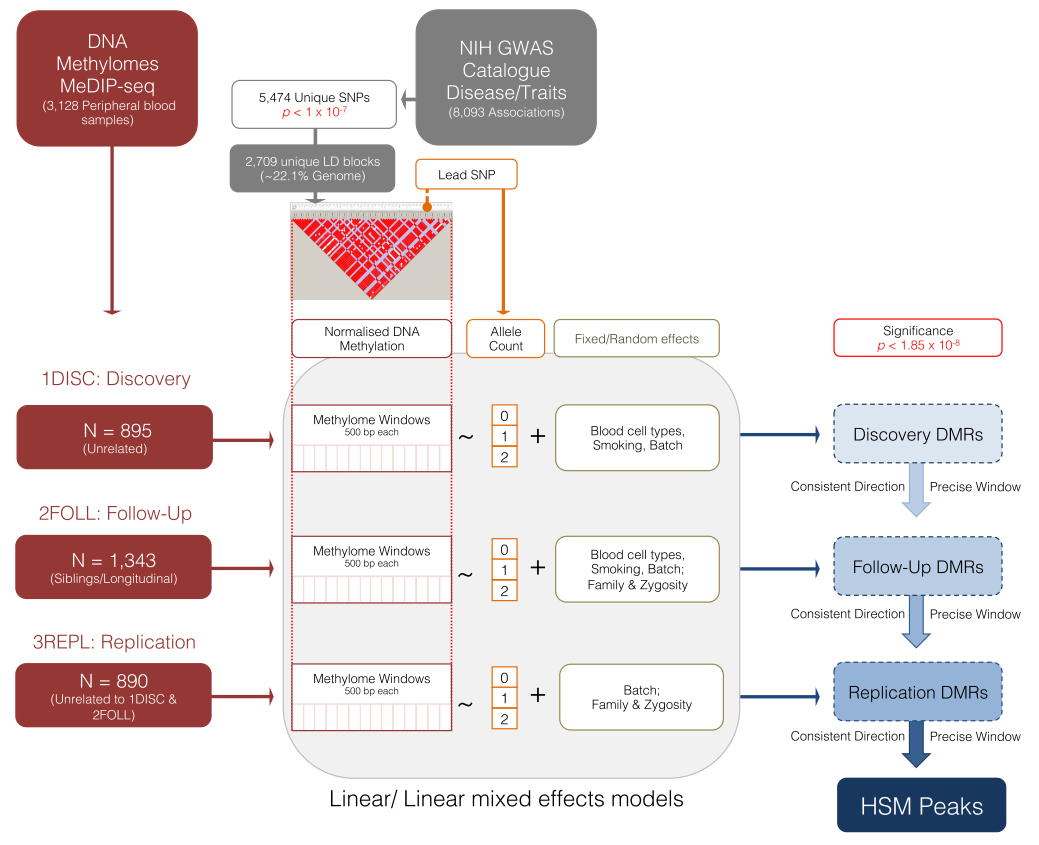
Please see the publication for more detailed discussion of these data and results.
This data is from MeDIP-seq experiments from the TwinsUK EpiTwin study.
Code for the Data analysis and plotting can be found at: github.com/epigenomed/hsm_analysis.
Code for this website can be found at: github.com/RichardJActon/hsm.